David N. Langelaan
Associate Professor

Email: david.langelaan@dal.ca
Phone: 902-494-8928
Mailing Address:
Sir Charles Tupper Medical Building
PO Box 15000
Halifax, Nova Scotia, Canada B3H 4R2
Education
- PhD, Dalhousie University
Academic Positions
- Department member since 2016
- Member of the Protein Assembly Research Team
Research Topics:
Structural biology, protein-protein interactions, cellular signalling, and protein engineering
Research
Are you interested in protein characterization and engineering?
We are! A large focus of my research group involves using biophysical techniques and functional assays to characterize the fundamental causes and consequences of protein assembly. To answer scientific questions we use techniques such as circular dichroism spectropolarimetry, isothermal titration calorimetry, nuclear magnetic resonance spectroscopy, X-ray diffraction, as well as functional assays using tissue culture. Three of our main research areas are described below. If you are interested in joining our team, please contact David Langelaan.
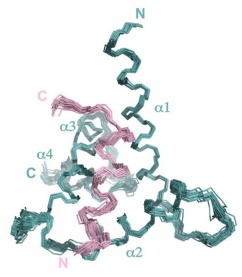
Hydrophobin characterization and engineering
Hydrophobins are small proteins that are ubiquitously expressed by filamentous fungi. These proteins are extremely surface active, form monolayers at interfaces and also structurally rearrange to become a functional amyloid that can be deposited onto surfaces. Due to their unique properties, hydrophobins have potential uses in industry. With the recent explosion of genomic data that is available, many new hydrophobin-coding sequences have been discovered that are not well classified or characterized. The goal of our lab is to gain mechanistic understanding into hydrophobin function by characterizing new hydrophobins and determining the mechanisms of their assembly. Furthermore, we are actively engineering hydrophobins to control their properties for commercial applications.
Deciphering the role of MITF in development and melanoma
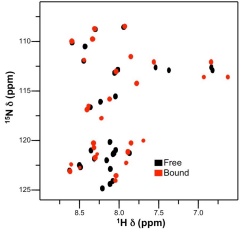
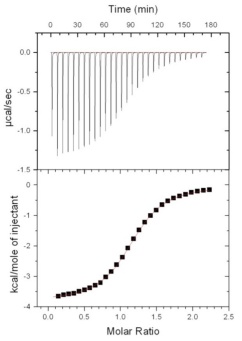
The microphthalmia family (MiT) of transcription factors are DNA-binding proteins that are responsible for recruiting transcription machinery and controlling expression of specific genes. They have important regulatory roles in a variety of cellular processes including pluripotency, homeostasis and cell differentiation. In particular, one member of the MiT family, MITF, is specifically expressed in melanocytes and is a master regulator of melanocyte development. Because of this critical role, mutation of MITF is implicated in diseases such as Waardenburg Syndrome type 2, Tietz syndrome and melanoma, where it serves as a lineage addiction oncogene. Our goal is to biophysically characterize the protein-protein interactions and post-translational modifications of MITF in both healthy and diseased states. This will allow us to gain an understanding of the mechanisms of MITF signalling and provide new routes of therapeutic intervention to treat MITF related diseases.
Characterizing the mechanism of Rhodoquinone biosynthesis
Life needs energy to survive. In organisms living in oxygen-rich environments, energy is obtained by aerobic respiration, a metabolic process requiring oxygen. Deprivation of oxygen severely damages many organisms, posing a serious threat to their survival. Some microbes have solved this problem by producing a molecule called rhodoquinone (RQ) that allows the respiratory system to make energy without oxygen. By ‘stealing’ the gene encoding RquA, an enzyme that produces RQ, these microbes have adapted to diverse low-oxygen environments distributed across the planet. Little is known about RquA, the chemical mechanism it employs to make RQ, and the impact of RQ on cellular respiration. We are using a multidisciplinary approach to determine the cellular, chemical, and structural aspects of RQ biosynthesis.
We thank the following agencies for providing funding to support our research:
Beatrice Hunter Cancer Research Institute, Cancer Research Training Program, CFI, NSERC, Nova Scotia Research and Innovation Trust, New Frontier in Research Fund, and the Dalhousie Medical Research Foundation
Keywords:
Gene cloning and mutagenesis, protein expression and purification, NMR spectroscopy, X-ray crystallography, isothermal titration calorimetry, fluorescence spectroscopy
Current Lab Members
| Alex Bouchard | Honours Student (2025) |
|---|---|
| Maegan Burke | Graduate Student (MSc) |
| Raymond He | Graduate Student (PhD) |
| Trilok Neupane | Graduate Student (PhD) |
| Dina Rogers | Graduate Student (MSc) |
| Janani Venkat | Honours Student (2025) |
| Amelia Wojtyk | Honours Student (2025) |
Publications
- Wijesundara NM, Lee SF, Langelaan DN, Lefsay A, Rupasinghe HPV (2024) Carvacrol alters the membrane phospholipid in erythromycin-resistant Streptococcus pyogenes. Phytomedicine Plus. 4:100614. DOI
- Brown AD, Lynch K, Langelaan DN (2023) The C-terminal transactivation domain of MITF interacts promiscuously with co-activator CBP/p300. Scientific Reports. 13:160994. DOI
- Brown AD, Vergunst KL, Branch M, Blair CM, Dupré DJ, Baillie G,Langelaan DN (2023) Structural basis of CBP/p300 recruitment by the microphthalmia-associated transcription factor. BBA-Mol Cell Res.1870:119520. DOI
- Grondin JM, Langelaan DN, Smith SP (2023) Qualitative and quantitative characterization of protein-carbohydrate interactions by NMR spectroscopy. Methods Mol Biol. 2657:115-128. DOI
- Brown AD, Cranstone C, Dupré DJ, Langelaan DN (2023) β-catenin interacts with the TAZ1 and TAZ2 domains of CBP/p300 to activate gene transcription. Int J Biol Macromol. 238:124155. DOI
- Mathavarajah S, Vergunst KL, Williams S, He R, Maliougina M, Habib EB, Park M, Salsman J, Roy S, Braasch I, Roger AJ, Langelaan DN*, Dellaire G* (*co-corresponding authors) (2023) PML and PML-like exonucleases restrict retrotransposons in jawed vertebrates. Nucleic Acids Res. 51:3185-3204. DOI
- Vergunst KL, Kenward C, Langelaan DN (2022) Characterization of the structure and self-assembly of two distinct class IB hydrophobins. Appl Microbiol Biotechnol. 106:7831-7843. DOI
- Wijesundara NM, Lee SF, Cheng Z, Davidson R, Langelaan DN, Rupasinghe HPV (2022) Bactericidal Activity of Carvacrol against Streptococcus pyogenes Involves Alteration of Membrane Fluidity and Integrity through Interaction with Membrane Phospholipids. Pharmaceutics. 14:1992. DOI
- Neupane T, Chambers LR, Godfrey AJ, Monlux MM, Jacobs EJ, Whitworth S, Spawn JE, Clingman SHK, Vergunst KL, Niven FM, Townley JJ, Orion IW, Goodspeed CR, Cooper KA, Cronk JD, Shepherd JN*, Langelaan DN* (*co-corresponding authors) (2022) Microbial rhodoquinone biosynthesis proceeds via an atypical RquA-catalyzed amino transfer from S-adenosyl-L-methionine to ubiquinone. Commun Chem. 5:89. https://doi.org/10.1038/s42004-022-00711-6
- Vergunst KL, Langelaan DN. (2022) The N-terminal tail of the hydrophobin SC16 is not required for rodlet formation. Sci Rep. 12:366 https://doi.org/10.1038/s41598-021-04223-6 [PubMed]
- Kenward C, Vergunst KL, Langelaan DN. (2020) Expression, purification, and refolding of diverse class IB hydrophobins. Protein Expr Purif. 176:105732. https://doi.org/10.1016/j.pep.2020.105732. PMID: 32866612.
- Salinas G, Langelaan DN, Shepherd JN. (2020) Rhodoquinone in bacteria and animals: Two distinct pathways for biosynthesis of this key electron transporter used in anaerobic bioenergetics. Biochim Biophys Acta Bioenerg. 1861(11):148278. https://doi.org/10.1016/j.bbabio.2020.148278. PMID: 32735860.
- Lochhead MR, Brown AD, Kirlin AC, Chitayat S, Munro K, Findlay JE, Baillie GS, LeBrun DP, Langelaan DN, Smith SP. (2020). Structural insights into TAZ2 domain-mediated CBP/p300 recruitment by transactivation domain 1 of the lymphopoietic transcription factor E2A. J Biol Chem. pii: jbc.RA119.011078. https://www.doi.org/10.1074/jbc.RA119.011078. [Epub ahead of print] [PubMed]
- Marczenko KM, Zurakowski JA, Kindervater MB, Jee S, Hynes T, Roberts N, Park S, Werner-Zwanziger U, Lumsden M, Langelaan DN, Chitnis SS. (2019). Periodicity in Structure, Bonding, and Reactivity for p-Block Complexes of a Geometry Constraining Triamide Ligand. Chemistry. 25(71):16414-16424. https://www.doi.org/10.1002/chem.201904361. [Epub 2019 Nov 26] [PubMed]
- Guo, S., Langelaan, D.N., Phippen, S.W., Smith, S.P., Voets, I.K., Davies, P.L., (2018) Conserved structural features anchor biofilm-associated RTX-adhesins to the outer membrane of bacteria FEBS J. 285(10):1812-1826 [PubMed] [Article]
- Langelaan, D.N., Pandey, A., Sarker, M. and Rainey, J.K., (2017) Preserved transmembrane segment topology, structure, and dynamics in disparate micellar environments J Phys Chem Lett 8:2381-2386 [PubMed] [Article]
- Grondin, J.M, Langelaan, D.N, Smith S.P., (2017) Quantifying protein-carbohydrate interactions by NMR. Methods in Molecular Biology, Wade Abbott Springer-Verlag, New York:143-156 [PubMed]
- Conroy, B.S., Weiss, E.R., Smith, S.P. and Langelaan, D.N., (2017) Backbone (1)H, (13)C, and (15)N NMR resonance assignments of the Krüppel-like factor 4 activation domain Biomol NMR Assign 11(1):95-98 [PubMed]
- Gandier, J.A., Langelaan, D.N., Won, A., O'Donnell, K., Grondin, J.L., Spencer, H.L., Wong, P., Tillier, E., Yip, C., Smith, S.P. and Master, E.R., (2017) Characterization of a Basidiomycota hydrophobin reveals the structural basis for a high-similarity Class I subdivision Sci. Rep. 10(7):45863 [PubMed]
- Guo, S., Stevens, C.A., Vance, T.D.R., Olijve, L.L.C., Graham, L.A., Campbell, R.L., Yazdi, S.R., Escobedo, C., Bar-Dolev, M., Yashunsky, V., Braslavsky, I., Langelaan, D.N., Smith, S.P., Allingham, J.S., Voets, I.K. and Davies, P.L. , (2017) Structure of a 1.5-MDa adhesin that binds its Antarctic bacterium to diatoms and ice Sci. Adv. 3(8):e1701440 [PubMed] [Article]
- Hettle, A., Fillo, A., Abe, K., Massel, P., Pluvinage, B., Langelaan, D.N., Smith, S.P. and Boraston, A.B., (2017) Properties of a family 56 carbohydrate-binding module and its role in the recognition and hydrolysis of β-1,3-glucan J. Biol. Chem. 292(41):16955-16968 [PubMed] [Article]
- Stevens, C.A., Semrau, J., Chiriac, D., Litschko, M., Campbell, R.L., Langelaan, D.N., Smith, S.P., Davies, P.L. and Allingham, J.S., (2017) Peptide backbone circularization enhances antifreeze protein thermostability Prot. Sci. 10:1932-1941 [PubMed] [Article]
- Ding Y., Nash J., Berezuk A., Khursigara C.M., Langelaan D.N., Smith S.P., and Jarrell K.F., (2016) Identification of the First Transcriptional Activator of an Archaellum Operon in a Euryarchaeon. Mol. Microbiol. 102:54-70 [PubMed]
- Langelaan, D.N., Liburd, J., Yang, Y., Miller, E., Chitayat, S., Crawley, S.W., Côté, G.P. and Smith, S.P. , (2016) Structure of the single-lobe myosin light chain C in complex with the light chain-binding domains of myosin-1C provides insights into divergent IQ-motif recognition J. Biol. Chem. 291:19607-17 [PubMed]
- Read J., Clancy E.K., Sarker M., de Antueno R., Langelaan D.N., Parmar H.B., Shin K., Rainey J.K., and Duncan R., (2015) Reovirus FAST Proteins Drive Pore Formation and Syncytiogenesis Using a Novel Helix-Loop-Helix Fusion-Inducing Lipid Packing Sensor. PLoS Pathog. 11(6): [PubMed]
- Denis C.M., Langelaan D.N., Kirlin A.C., Chitayat S., Munro K., Spencer H.L., LeBrun D.P., and Smith S.P., (2014) Functional redundancy between the transcriptional activation domains of E2A is mediated by binding to the KIX domain of CBP/p300. Nucleic Acids. Res. 42(11):7370-7382 [PubMed]
- Langelaan, D.N., Reddy, T., Banks, A.W., Dellaire, G., Dupré, D. and Rainey, J.K., (2013) Structural features of the apelin receptor N-terminal tail and first transmembrane segment implicated in ligand binding and receptor trafficking. Biochimica et Biophysica Acta - Biomembranes 1828:1471-1483 [PubMed] [Article]
- Langelaan, D.N.*, Ngweniform, P.*, Rainey, J.K. (*contributed equally), (2011) Biophysical characterization of G-protein coupled receptor-peptide ligand binding. Biochem Cell Biol 89:98-105 [PubMed]
- Langelaan, D.N. and Rainey, J.K., (2010) Membrane catalysis of peptide-receptor binding. Biochem Cell Biol 88:203-210 [PubMed]
- Langelaan, D.N., Wieczorek, M., Blouin, C. and Rainey, J.K., (2010) Improved helix and kink characterization in membrane proteins allows evaluation of kink sequence predictors. J. Chem. Inf. Model. 50:2213-2220 [PubMed]
- Langelaan, D.N., Bebbington, E.M., Reddy, T. and Rainey, J.K., (2009) Structural insight into G-protein coupled receptor binding by apelin Biochemistry 48:537-548 [PubMed]
- Langelaan, D.N. and Rainey, J.K., (2009) Headgroup-dependent membrane catalysis of apelin-receptor interactions is likely. J Phys Chem B 113:10465-10471 [PubMed]