Graham Dellaire
Professor

Email: dellaire@dal.ca
Phone: 902-494-4730
Fax: 902-494-2519
Mailing Address:
Sir Charles Tupper Medical Building
PO Box 15000
Halifax, Nova Scotia, Canada B3H 4R2
Education
- PhD, McGill University
Academic Positions
- Department member since 2007
- Director of Research, Pathology
Research Topics
Nuclear structure and cancer biology; chemotherapy resistance and cancer biomarkers; CRISPR genome engineering; the role of nuclear organization in DNA repair, tumour suppression, and cell cycle control; genome instability and pre-mRNA splicing; zebrafish as a novel animal tumour model.
Research
Nuclear Structure and Cancer Biology
Genomic instability contributes to both the development and progression of cancer. Mutations affecting the detection or repair of DNA damage can dramatically increase genome instability and therefore cancer susceptibility.
A key question in cancer biology is how the cell detects and repairs DNA damage?
We are studying how the structure of the mammalian nucleus, including chromatin and the various nuclear domains, contributes to DNA damage surveillance, signaling and repair.
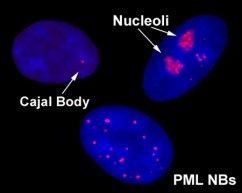
Promyelocytic leukemia nuclear bodies (PML NBs): novel biomarkers for the cellular response to DNA damage.
Finding and repairing damaged DNA among the 3 billion base pairs of the human genome is an astonishing cellular feat, the complexity of which we are only beginning to appreciate. DNA in human cells exists as a nucleoprotein complex known as chromatin, which is in turn compacted and folded into individual chromosomes that form distinct territories in the nucleus. It is within this context that DNA damage must be detected and repaired. An early event in signaling DNA damage is the modification of chromatin at a DNA double-strand break (DSB) by phosphorylation of the histone variant H2A.X by the kinases ATM, ATR and DNA-PK. DNA damage also triggers the global decondensation of chromatin, mediated partly by the ATM kinase, as well as the structural reorganisation of several subnuclear compartments including nucleoli, Cajal bodies and promyelocytic leukemia nuclear bodies (PML NBs)(Dellaire and Bazett-Jones, 2004).
In my laboratory, we are interested in characterizing how DNA damage affects the structure and function of the mammalian nucleus with the hope of determining signature changes in nuclear architecture that can be used as biomarkers for cancer diagnosis, staging and treatment. We use a combination of biochemistry, molecular biology and molecular imaging approaches to determine how changes in nuclear structure contribute to DNA repair.
Recent work from my laboratory has identified the PML NB as a possible DNA damage sensor, which can respond to ionizing radiation and chemotherapy treatments (Dellaire and Bazett-Jones, 2004; Dellaire et al., 2006). Currently, we are addressing how PML NBs may contribute to DNA damage surveillance, signaling and repair. These studies will provide needed insight into the role of these domains in tumour suppression and may provide a useful paradigm for the study of the role of other nuclear domains in oncogenesis.
Xenotransplantation in zebrafish as a human tumour model
Relevant human tumour models are needed more than ever to accelerate the translation of basic cancer research to the clinic. In collaboration with pediatric oncologist Dr. Jason Berman (Dept. of Pediatrics, Dalhousie University and IWK Hospital, Halifax) we have been developing the novel human tumour animal model based on the xenotransplantation of human tumour cells in the zebrafish embryo.
Current applications of this tumour model include:
- The study of chemotherapy resistance in breast cancer (Breast Cancer Society/QEII Foundation)
- Zebrafish as a model for evaluating new treatments, their toxicity, and affect on metastasis in prostate cancer (Prostate Cancer Canada) DalNews Story on this work
- The study of tumour metastasis in pediatric sarcomas (C17 Council) - completed
- Drug screening for novel chemotherapeutics targeting leukemia (CIHR/NSERC Collaborative Health Research Program) - completed Global News Story on this work
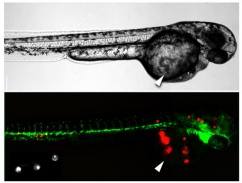
Twenty four hour post fertilization Fli-GFP transgenic zebrafish embryos expressing the green fluorescent protein to highlight their blood vessels were microinjected with 50 leukemia cells (labeled in red) within the yolk sac (white arrow). A zebrafish embryo is shown by phase contrast (top panel) and fluorescence microscopy (bottom panel) at 24 hours post-injection. Red fluorescent masses of leukemia cells can be seen in the tail and head of the embryo indicating that the leukemia cells can enter the blood stream and migrate via the vasculature to distant sites from the injection site.
Enhancing CRISPR Genome Engineering
In our laboratory, we have been working on methods to enhance gene editing by CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats), a primordial immune system found in many forms of bacteria that can cut DNA in a site-specific manner using small guide RNAs (sgRNAs). Although many research groups are actively pursuing genetic approaches to enhancing CRISPR-based gene editing, these approaches are not very efficient and require knocking-out important genes in DNA repair; an approach that is less than ideal in terms of safety for patients undergoing gene therapy using CRISPR. Therefore, we have focused on the discovery of small molecules that could transiently (and reversibly) modulate DNA repair to enhance gene editing. Towards this goal, we developed a small molecule screen based on the insertion of the bright EGFP variant gene Clover into the lamin A/C locus (LMNA) by CRISPR-mediated HDR (Pinder et al., 2015) and this system was used in collaboration with the group of Dr. Daniel Durocher to identify the molecular basis for the inhibition of HR in non-dividing cells, an advance that could open the way to gene therapy of diseases such as muscular dystrophies that affect non-dividing muscle cells (Orthwein et al., 2015).
Our CRISPR/Cas9 studies have received considerable media attention, see:
- 2016 Global News Story LINK
- Cover Story, 2016 Spring Issue of the Philanthropist LINK
We continue to develop novel assays and tools using CRISPR technology for the study of DNA repair as well as develop novel therapies for cancer and genetic diseases.
Our current projects include:
- Enhancing the efficiency of CRISPR-mediated Homology Directed Repair (HDR) genetically and through novel small molecules
- Applying CRISPR-HDR to the identification and characterization of nuclear subdomains and their associated proteins
- Development of novel transgenic and gene-modified zebrafish for the study of cancer (in collaboration with Dr. Jason Berman (Dept. of Microbiology, Dalhousie University).
More more details and a brief history of gene editing and CRISPR, please follow this LINK
Keywords:
cancer biology, DNA repair, CRISPR gene editing, biomarkers, molecular and cellular imaging, zebrafish
Current Lab Members
| Joyce Chew |
Lab Technician |
|---|---|
| Livia Clarke |
Grad Student (MSc, Pathology) |
| Jayme Salsman |
Postdoc (Dalhousie University) |
| Carter Van Iderstine | Grad Student (MSc, Pathology) (co-supervised with K. Bedard) |
Publications
- Tam, N.W., Chung, D., Baldwin, S.J., Simmons, J.R., Xu, L., Rainey, J.K., Dellaire, G., and Frampton, J.P. (2020) Material properties of disulfide-crosslinked hyaluronic acid hydrogels influence prostate cancer cell growth and metabolism. J. Mater. Chem. B. 8: 9718-9733. [PubMed] [Article]
- Corkery DP, Clarke LE, Gebremeskel S, Salsman J, Pinder J, Le Page C, Meunier L, Xu Z, Mes-Masson AM, Berman JN, Johnston B, Dellaire G, (2018) Loss of PRP4K drives anoikis resistance in part by dysregulation of epidermal growth factor receptor endosomal trafficking. Oncogene in press:doi: 10.1038/onc.2017.318 [PubMed] [Article]
- Salsman J, Masson JY, Orthwein A, Dellaire G., (2018) CRISPR/Cas9 Gene Editing: From Basic Mechanisms To Improved Strategies For Enhanced Genome Engineering In Vivo. Current Gene Therapy in press DOI: 10.2174/1566523217666171122094629: [PubMed] [Article]
- Pauty, J., A. Couturier,A. Rodrigue, M. Caron, Y. Coulombe. G. Dellaire and J.-Y. Masson , (2017) Cancer-causing mutations in the tumour suppressor PALB2 reveals a novel cancer mechanism using hidden nuclear export signal in the WD40 repeat motif Nucleic Acids Research 45(5):2644-2657 [PubMed]
- Salsman, J. and G. Dellaire , (2017) Precision Genome Editing in the CRISPR Era Biochemistry & Cell Biology 95(2):187-201 [PubMed] [Article]
- Rémi Buisson, Joshi Niraj, Amélie Rodrigue, Chu Kwen Ho, Johannes Kreuzer, Tzeh Keong Foo, Emilie J.-L. Hardy, Graham Dellaire, Wilhelm Haas, Bing Xia, Jean-Yves Masson, Lee Zou, (2017) Coupling of Homologous Recombination and the Checkpoint by ATR Molecular Cell 65(2):336-346 [PubMed] [Article]
- Salsman J, Stathakis A, Parker E, Chung D, Anthes LE, Koskowich KL, Lahsaee S, Gaston D, Kukurba KR, Smith KS, Chute IC, Léger D, Frost LD, Montgomery SB, Lewis SM, Eskiw C, Dellaire G., (2017) PML nuclear bodies contribute to the basal expression of the mTOR inhibitor DDIT4 Scientific Reports 7:45038 [PubMed] [Article]
- Salsman J, Rapkin LM, Margam NN, Duncan R, Bazett-Jones DP, Dellaire G, (2017) Myogenic differentiation triggers PML nuclear body loss and DAXX relocalization to chromocentres Cell Death and Disease 8(3):e2724 [PubMed] [Article]
- Agudelo D, Duringer A, Bozoyan L, Huard CC, Carter S, Loehr J, Synodinou D, Drouin M, Salsman J, Dellaire G, Laganière J, Doyon Y., (2017) Marker-free coselection for CRISPR-driven genome editing in human cells. Nature Methods 14(6):615-620 [PubMed] [Article]
- Lobbardi R, Pinder J, Martinez-Pastor B, Theodorou M, Blackburn JS, Abraham BJ, Namiki Y, Mansour M, Abdelfattah NS, Molodtsov A, Alexe G, Toiber D, de Waard M, Jain E, Boukhali M, Lion M, Bhere D, Shah K, Gutierrez A, Stegmaier K, Silverman LB, Sadreyev RI, Asara JM, Oettinger MA, Haas W, Look AT, Young RA, Mostoslavsky R, Dellaire G, Langenau DM., (2017) TOX Regulates Growth, DNA Repair, and Genomic Instability in T-cell Acute Lymphoblastic Leukemia. Cancer Discovery 7(11):1336-1353 [PubMed] [Article]
- Gebremeskel S, Lobert L, Tanner K, Walker B, Oliphant T, Clarke LE, Dellaire G, Johnston B., (2017) Natural Killer T-cell Immunotherapy in Combination with Chemotherapy-Induced Immunogenic Cell Death Targets Metastatic Breast Cancer. Cancer Immunol. Res. 5(12):1086-1097 [PubMed] [Article]
- Melong N, Steele S, MacDonald M, Holly A, Collins CC, Zoubeidi A, Berman JN, Dellaire G., (2017) Enzalutamide inhibits testosterone-induced growth of human prostate cancer xenografts in zebrafish and can induce bradycardia. Scientific Reports 7(1):14698. [PubMed] [Article]
- Lahsaee S, Corkery DP, Anthes LE, Holly A, Dellaire G., (2016) Estrogen receptor alpha (ESR1)-signaling regulates the expression of the taxane-response biomarker PRP4K. Experimental Cell Research 340(1):125-31. [PubMed] [Article]
- Khan IA, Yoo BH, Masson O, Baron S, Corkery D, Dellaire G, Attardi LD, Rosen KV., (2016) ErbB2-dependent downregulation of a pro-apoptotic protein Perp is required for oncogenic transformation of breast epithelial cells. Oncogene 35(44):5759-5769. [PubMed]
- Rajan V, Dellaire G, Berman JN., (2016) Modeling Leukemogenesis in the Zebrafish Using Genetic and Xenograft Models. Methods Mol Biol. 1451:171-89 [PubMed]
- Wertman J, Veinotte CJ, Dellaire G, Berman JN., (2016) The Zebrafish Xenograft Platform: Evolution of a Novel Cancer Model and Preclinical Screening Tool. Adv Exp Med Biol. 916:289-314 [PubMed]
- George VC, Dellaire G, Rupasinghe HP., (2016) Plant flavonoids in cancer chemoprevention: role in genome stability. Journal of Nutritional Biochemistry 45:1-14 [PubMed] [Article]
- Couturier AM, Fleury H, Patenaude AM, Bentley VL, Rodrigue A, Coulombe Y, Niraj J, Pauty J, Berman JN, Dellaire G, Di Noia JM, Mes-Masson AM, Masson JY., (2016) Roles for APRIN (PDS5B) in homologous recombination and in ovarian cancer prediction. Nucleic Acids Research 44(22):10879-10897. [PubMed] [Article]
- Bentley, V., C. Veinotte, D. Corkery, J. Pinder, M.A. Leblanc, K. Bedard, A.P Weng, J.N. Berman, and G. Dellaire, (2015) Focused chemical genomics using zebrafish xenotransplantation as a preclinical therapeutic platform for T-cell acute lymphoblastic leukemia Haematologica 100(1):70-6 [PubMed] [Article]
- Sidik, S., J. Salsman, G. Dellaire, and J. Rohde, (2015) Shigella infection interferes with SUMOylation and increases PML-NB number PLoS ONE 10(4):e0122585 [PubMed] [Article]
- Kalousi, A., A-S. Hoffbeck, P. Selemenakis, J. Pinder, K.I. Savage, K.K. Khanna, L. Brino, G. Dellaire, V.G. Gorgoulis, and E. Soutoglou , (2015) The nuclear oncogene SET controls DNA repair by KAP1 and HP1 retention on chromatin Cell Reports 11(1):149-63 [PubMed] [Article]
- Corkery, D.P., L. Meunier , C. Le Page , D.M. Provencher , A-M Mes-Masson and G. Dellaire, (2015) PRP4K is a novel Her2-regulated biomarker of taxane sensitivity Cell Cycle 14(7):1059-69 [PubMed] [Article]
- El-Naggar, A.M., C.J Veinotte, H. Cheng, T.G.P. Grunewald, G. Luca Negri, S. Prakash Somasekharan, D.P. Corkery, F. Tirode, J. Mathers, D. Khan, A.H Kyle, J.H Baker, N.E. LePard, S.McKinney, S.Hajee, M. Bosiljcic, G. Leprivier, C.E. Tognon, A.I. Minchinton, K.L. Bennewith, O.Delattre, Y. Wang, G. Dellaire, J.N. Berman, and P.H. Sorensen., (2015) Tranlsational activation of HIF1a by YB-1 promotes sarcoma metastasis Cancer Cell 27(5):682-97 [PubMed] [Article]
- Corkery, D., A. Holly, S. Lahsaee, and G. Dellaire, (2015) Connecting the Speckles: Splicing kinases and their role in tumorigenesis and treatment response Nucleus 6(4):279-88 [PubMed] [Article]
- Chung, D., and G. Dellaire. , (2015) The role of the COP9 signalosome and neddylation in DNA damage signaling and repair Biomolecules in press:
- Pinder, J., J. Salsman, and G. Dellaire, (2015) Nuclear domain knock-in screen for the evaluation and identification of small molecule enhancers of CRISPR-based genome editing Nucleic Acids Research 43(19):9379-92 [PubMed] [Article]
- Corkery DP, Le Page C, Meunier L, Provencher D, Mes-Masson AM, Dellaire G., (2015) PRP4K is a HER2-regulated modifier of taxane sensitivity Cell Cycle 14(7):1059-69 [PubMed] [Article]
- Kalousi A, Hoffbeck AS, Selemenakis PN, Pinder J, Savage KI, Khanna KK, Brino L, Dellaire G, Gorgoulis VG, Soutoglou E., (2015) The nuclear oncogene SET controls DNA repair by KAP1 and HP1 retention to chromatin. Cell Reports 11(1):149-63. [PubMed] [Article]
- Sidik SM, Salsman J, Dellaire G, Rohde JR., (2015) Shigella infection interferes with SUMOylation and increases PML-NB number. PLoS ONE 10(4):e0122585 [PubMed] [Article]
- El-Naggar AM, Veinotte CJ, Cheng H, Grunewald TG, Negri GL, Somasekharan SP, Corkery DP, Tirode F, Mathers J, Khan D, Kyle AH, Baker JH, LePard NE, McKinney S, Hajee S, Bosiljcic M, Leprivier G, Tognon CE, Minchinton AI, Bennewith KL, Delattre O, Wang Y, Dellaire G, Berman JN, Sorensen PH., (2015) Translational Activation of HIF1α by YB-1 Promotes Sarcoma Metastasis. Cancer Cell 27(5):682-97 [PubMed] [Article]
- Corkery DP, Holly AC, Lahsaee S, Dellaire G., (2015) Connecting the speckles: Splicing kinases and their role in tumorigenesis and treatment response. Nucleus 6(4):279-88 [PubMed] [Article]
- Orthwein A, Noordermeer SM, Wilson MD, Landry S, Enchev RI, Sherker A, Munro M, Pinder J, Salsman J, Dellaire G, Xia B, Peter M, Durocher D., (2015) A mechanism for the suppression of homologous recombination in G1 cells. Nature 528(7582):422-6 [PubMed] [Article]
- Stairs CW, Eme L, Brown MW, Mutsaers C, Susko E, Dellaire G, Soanes DM, van der Giezen M, Roger AJ., (2014) A SUF Fe-S Cluster Biogenesis System in the Mitochondrion-Related Organelles of the Anaerobic Protist Pygsuia Curr. Biol 24(11):1176-1186 [PubMed] [Article]
- Liu, Y., A. Asnani, L. Zou, V.L. Bentley, M. Yu, Y.Wang, G.Dellaire, K.S. Sarkar, M. Dai, H.H. Chen, D.E. Sosnovik, J.T. Shin, D.A. Haber, J.N. Berman, W. Chao, and R.T. Peterson , (2014) Visnagin protects against doxorubicin-induced cardiomyopathy through modulation of mitochondrial malate dehydrogenase Science Translational Medicine 6(266):266ra170 [PubMed] [Article]
- Lemaître, C., A. Grabarz, K.Tsouroula, L. Andronov, A. Furst, T. Pankotai, V.Heyer, M. Rogier, K.M. Attwood, P. Kessler, G. Dellaire, B. Reina-San-Martin, and E. Soutoglou , (2014) Nuclear position dictates DNA repair pathway choice Genes & Development 28(22):2450-63 [PubMed] [Article]
- Klement, K., M.S. Luijsterburg, J. Pinder, C.S. Cena, V. Del Nero1, C.M. Wintersinger, G. Dellaire, H. van Attikum, A.A. Goodarzi, (2014) Opposing ISWI- and CHD-class chromatin remodeling activities orchestrate heterochromatic DNA repair Journal of Cell Biology 207(6):717-733 [PubMed] [Article]
- Smithen DA, Forrester AM, Corkery DP, Dellaire G, Colpitts J, McFarland SA, Berman JN, Thompson A., (2013) Investigations regarding the utility of prodigiosenes to treat leukemia. Org. Biomol. Chem. 11(1):62-8 [PubMed]
- Langelaan, D.N., Reddy, T., Banks, A.W., Dellaire, G., Dupré, D. and Rainey, J.K., (2013) Structural features of the apelin receptor N-terminal tail and first transmembrane segment implicated in ligand binding and receptor trafficking. Biochimica et Biophysica Acta - Biomembranes 1828:1471-1483 [PubMed] [Article]
- Pinder JB, Attwood KM, Dellaire G., (2013) Reading, writing, and repair: the role of ubiquitin and the ubiquitin-like proteins in DNA damage signaling and repair. Front Genet. 4:45 [PubMed]
- Andrin C, McDonald D, Attwood KM, Rodrigue A, Ghosh S, Mirzayans R, Masson JY, Dellaire G, Hendzel MJ., (2012) A requirement for polymerized actin in DNA double-strand break repair. Nucleus 3(4):384-95 [PubMed]
- Salsman J, Jagannathan M, Paladino P, Chan PK, Dellaire G, Raught B, Frappier L., (2012) Proteomic profiling of the human cytomegalovirus UL35 gene products reveals a role for UL35 in the DNA repair response. J. Virol. 86(2):806-20. [PubMed]
- Konantz M, Balci TB, Hartwig UF, Dellaire G, André MC, Berman JN, Lengerke C., (2012) Zebrafish xenografts as a tool for in vivo studies on human cancer. Ann N Y Acad Sci. 1266:124-37 [PubMed]
- Kepkay, R., Attwood, K.M., Ziv, Y., Shiloh, Y., Dellaire G., (2011) KAP1 depletion increases PML nuclear body number in concert with ultrastructural changes in chromatin. Cell Cycle 10(2):308-222 [PubMed]
- Cann KL, Dellaire G., (2011) Heterochromatin and the DNA damage response: the need to relax Biochem Cell Biol. 89(1):45-60 [PubMed]
- Corkery DP, *Dellaire G, *Berman JN., (2011) Leukaemia xenotransplantation in zebrafish--chemotherapy response assay in vivo. *Co-corresponding authors Brit. J. Haematol. 153(6):786-9 [PubMed]
- Bauer DC, Willadsen K, Buske FA, Lê Cao KA, Bailey TL, Dellaire G, Bodén M., (2011) Sorting the nuclear proteome. Bioinformatics 27(13):i7-14. [PubMed]
- Dellaire, G., Kepkay, R., Bazett-Jones, D.P., (2009) High resolution imaging of changes in the structure and spatial organization of chromatin, gamma-H2A.X and the MRN complex within etoposide-induced DNA repair foci. Cell Cycle 8(22):3750-3769 [PubMed]
- Dellaire, G. and D.P. Bazett-Jones, (2007) Beyond Repair Foci: Subnuclear Domains and the Cellular Response to DNA Damage Cell Cycle. 6(15):1864-1872 [PubMed]
- Dellaire, G., R. Ching, K. Ahmed, K. Tse, F. Jalali, R.G. Bristow, and D.P. Bazett-Jones, (2006) PML nuclear bodies are DNA damage sensors whose response to DNA double-strand breaks is regulated by NBS1, Chk2 and ATR J. Cell Biol. 175(1):55-66 [PubMed]
- Dellaire, G., R. Ching, H. Dehghani, Y. Ren and D.P. Bazett-Jones, (2006) PML nuclear bodies increase in number in early S-phase by a fission mechanism J. Cell Sci. 119(6):1026-1033 [PubMed]
- Dellaire, G., C. Eskiw, H. Dehghani, R. Ching, and D.P. Bazett-Jones, (2006) Mitotic Accumulations of PML protein (MAPPs) contribute to the post-mitotic re-establishment of PML nuclear bodies. J. Cell Sci. 119(6):1034-1042 [PubMed]
- Kruhlak, M.J., A. Celeste, G. Dellaire, O. Fernández-Capetillo,W.G. Müller, J.G. McNally, D.P. Bazett-Jones, and A. Nussenzweig, (2006) Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks J. Cell Biol. 172(6):823-834 [PubMed]